Oral Abstract
Automated Quantification of Left Ventricular Scar Volume in Cardiac MRI using Large Vision Models
- NT
Neda Tavakoli, PhD
Postdoctoral Fellow
Northwestern University - NT
Neda Tavakoli, PhD
Postdoctoral Fellow
Northwestern University 
Daniel Lee, MD, MSc
Professor of Medicine and Radiology
Northwestern University Feinberg School of Medicine
Northwestern- AR
Amir Ali Rahsepar, MD
Assistant Professor
Northwestern University - BB
Brandon Benefield, MSc
Research Lab Manager
Northwestern University 
Daming Shen, MSc, BSc
PhD
Northwestern University- SL
Santiago López-Tapia, PhD
postdoctoral fellow
Northwestern University - FS
Florian Schiffers, MSc
PhD Candidate
Northwestern University - EW
Edwin Wu, MD
Cardiologist
Northwestern University - AK
Aggelos Katsaggelos, PhD
Professor
Northwestern University
Presenting Author(s)
Primary Author(s)
Co-Author(s)
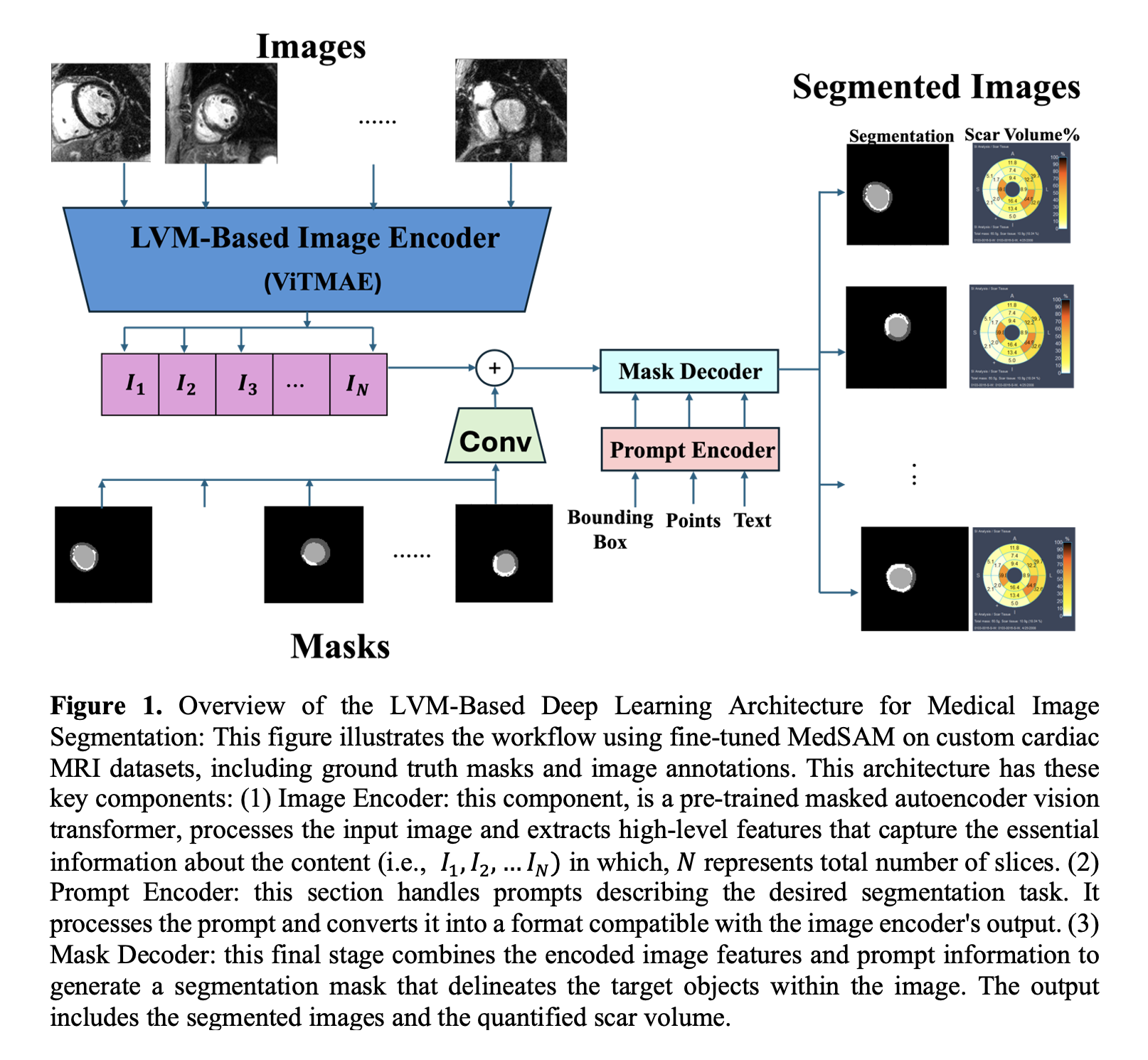
Late gadolinium enhancement (LGE) is the gold standard for evaluating myocardial fibrosis and scarring. Despite its importance, LGE-derived left ventricular (LV) scar volume is underutilized in clinical practice due to the time-consuming manual segmentation process and its susceptibility to inter-observer variability. Initial attempts to automate this process using deep learning (DL) models have shown limited success. A 2021 meta-analysis reported suboptimal Dice scores for LV scar quantification, with a mean Dice score (DSC) of 0.616 for supervised and 0.633 for unsupervised architectures. Figure 1 illustrates the proposed method leveraging large vision models (LVMs) to address these challenges.
Methods:
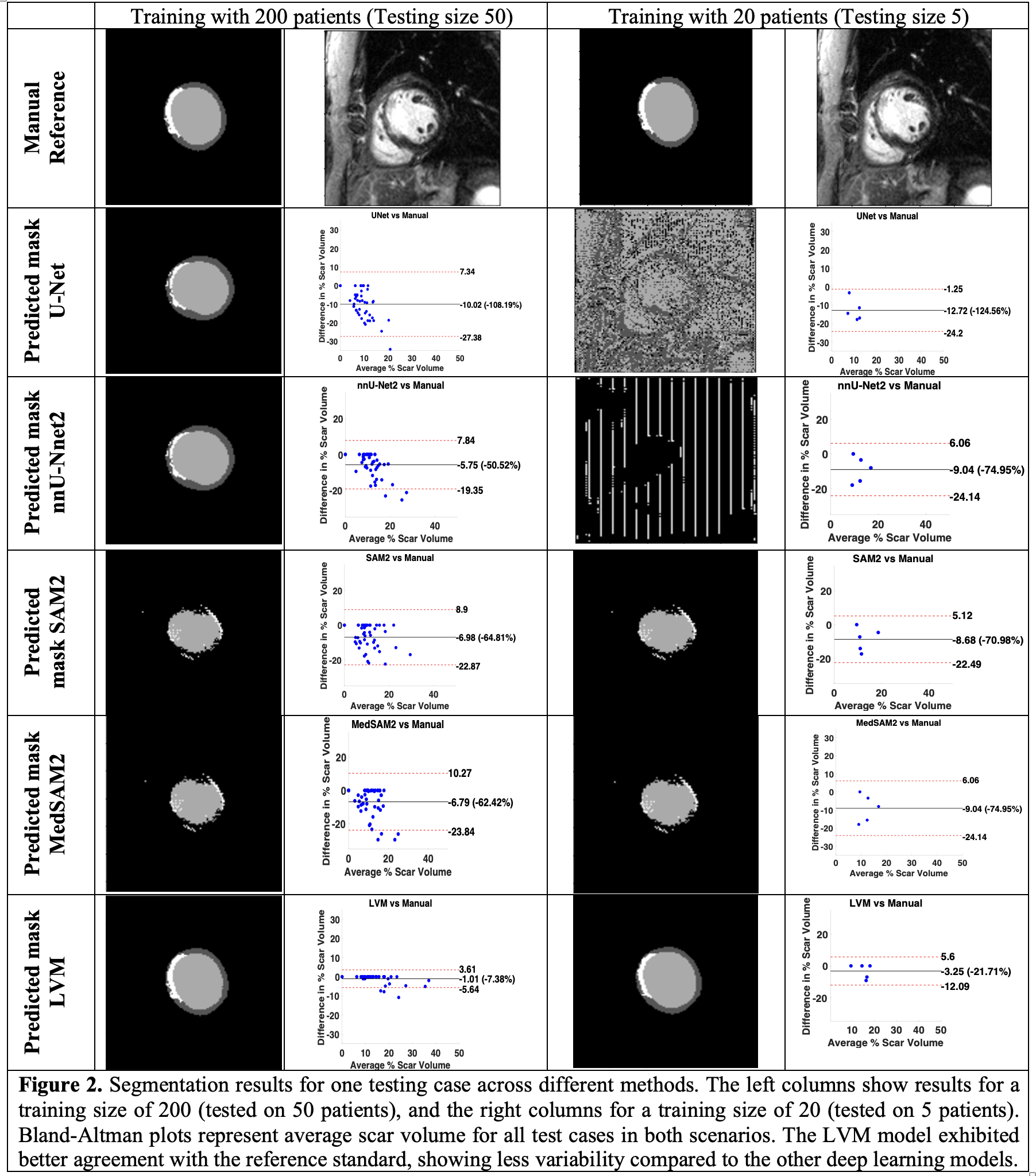
We utilized LGE data from the DETERMINE Trial registry (ClinicalTrials.gov ID NCT00487279), focusing on patients with ischemic cardiomyopathy and LVEF < 50%. A fine-tuned version of the MedSAM model was employed to segment LV into four classes: background, healthy myocardium, scar tissue, and LV blood pool. Training and testing were performed on GPU workstations using PyTorch. The dataset consisted of 2445 2D images for training and 437 images for testing. We evaluated performance across varying training and testing sizes, analyzing the impact on Dice scores. Figure 2 provides representative segmentation results comparing models and training sizes. The performance of our LVM-based model was compared to U-Net, nnU-Net2, SAM2, and MedSAM2 models.
Results:
Figure 2 shows representative segmentation results comparing models trained with 200 versus 20 patients. Models like U-Net and nnU-Net2, which learn from scratch, perform poorly with smaller training sizes, while pre-trained models like SAM2, MedSAM2, and our LVM perform better. Our LVM, pre-trained on MedSAM and fine-tuned with a custom LGE dataset, outperforms others in LV scar segmentation, especially with 200 patients. Compared to manual segmentation, for a training size of 200 patients, our LVM-based model (bias = -1.01% [-7.38% of mean]; CV = -232.63%) showed better agreement than U-Net (bias = -10.02% [-108.19% of mean]; CV = -88.41%), nnU-Net2 (bias = -5.75% [-50.52% of mean]; CV = -120.55%), SAM2 (bias = -8.68% [-70.98% of mean]; CV = -116.04%), and MedSAM2 (bias = -6.79% [-62.42% of mean]; CV = -128.18%), with lower bias and narrower limits of agreement.
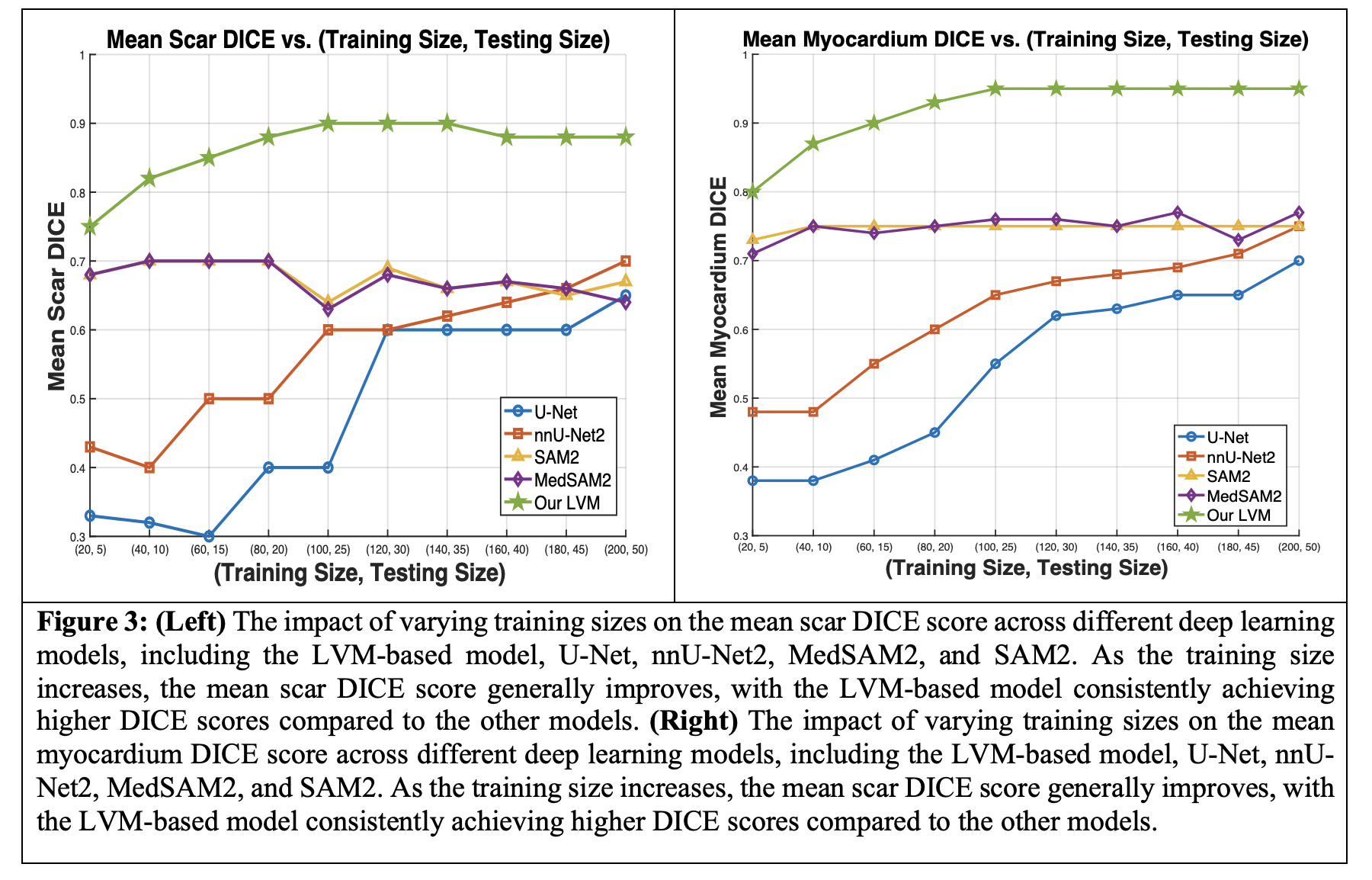
Figure 3 (Left) illustrates the effect of varying training sizes on mean scar DICE scores. SAM2 and MedSAM2 show little change, while U-Net and nnU-Net2 improve with more data, though U-Net plateaus. Our LVM model starts with a higher DICE score and remains stable as training data increases. Figure 3 (Right) shows smoother improvement in myocardium DICE scores across models, with larger myocardium structures being easier to segment. SAM2 and MedSAM2 show no significant improvement without additional training.
Conclusion:
Our fine-tuned LVM model outperforms existing DL models in LV scar volume quantification, achieving higher DICE scores and better agreement with manual measurements. For 200 patients, our LVM showed no significant difference from manual measurements for both scar volume (median = 0.123 [IQR = 0.084]; p > 0.999) and myocardial mass (median = 0.927 [IQR = 0.090]; p > 0.999), unlike U-Net, SAM2, and MedSAM2, which differed significantly. This success, driven by extensive pre-training and fine-tuning, makes our LVM a more reliable method for LV scar quantification, crucial for predicting adverse cardiac events and improving patient care.