Science Sessions
Predicting Hemorrhagic Myocardial Infarction in Patients Prior to Reperfusion with Explainable AI
- KY
Khalid Youssef, PhD
Assistant Research Professor
Indiana University, Department of Radiology and Imaging Sciences - KY
Khalid Youssef, PhD
Assistant Research Professor
Indiana University, Department of Radiology and Imaging Sciences 
Keyur P. Vora, MD, MS, FACC
Cardiologist, Assistant Professor of Medicine
Indiana University School of Medicine- RG
Rajesh Gupta, MD
Assistant Professor
University of Toledo 
Rohan Dharmakumar, PhD
Executive Director
Indiana University School of Medicine
Presenting Author(s)
Primary Author(s)
Co-Author(s)
Intramyocardial Hemorrhage (IMH) is a severe complication following reperfusion therapy for ST-segment elevation myocardial infarction (STEMI). IMH occurs in approximately 40% of successfully reperfused STEMI patients, significantly elevating the risk of mortality, major adverse cardiovascular events (MACE), and heart failure hospitalizations. T2* CMR is the gold standard for diagnosing IMH. Among the many challenges (access to CMR in acute setting and cost), T2* can only identify hemorrhagic MI (hMI) in patients after reperfusion. If hMI could be predicted prior to reperfusion, it would open opportunities to mitigate the development of hMI or its effects in a timely manner.
We tested a novel predictive algorithm for identifying patients at risk of IMH using explainable AI (XAI) based on key clinical predictors readily available during emergency coronary angiography. Unlike traditional "black box" deep learning models that provide predictions without clear reasoning, XAI ensures transparency by revealing how each feature contributes to IMH risk. This approach makes the model both accurate and interpretable in the clinical environment, resulting in a user-friendly point system that enables real-time, informed decision-making during PCI to potentially improve patient outcomes.
Methods:
STEMI patients (n=264) who underwent percutaneous coronary intervention (PCI) were prospectively enrolled, with CMR and T2* mapping preformed within 48-72 hours post-PCI to establish IMH ground truth (ClinicalTrials.gov ID NCT06423625). Patients were selected based on the availability of complete clinical data, including pre-PCI ECG and coronary angiogram, and post-PCI CMR. Demographics, medical history, and relevant clinical variables were also recorded for data analysis. The Superposable Neural Network (SNN) XAI method was employed to identify the best predictors of IMH, ensuring accuracy against T2* CMR while creating a transparent model that clearly illustrates how each predictor relates to the probability of developing IMH.
Results:
A representative set of pre-PCI coronary angiograms and corresponding T2* CMR from two patients (one with IMH and another patient without IMH) are shown in Fig. 1. Probability of developing IMH and its relation to the various predictors is shown in Fig. 2. From the explanations provided by the SNN model, a simplified point system was developed (Fig. 3).
Our findings here support the notion that the presence of collaterals, degree of occlusion, and ST-segment score (based on extent of ST elevation) can predict IMH with an AUC of 92%. An easily usable point system as shown in Fig. 3, demonstrated an accuracy of 84.9%, sensitivity of 82.3% sensitivity, specificity of 87.3% for predicting IMH prior to reperfusion.
Conclusion: ST-elevation MI patients who are diagnosed for IMH with CMR days after reperfusion therapy can be identified prior to reperfusion from key predictors distilled from explainable AI based on Superposable Neural Network (SNN) with high accuracy and transparency. The simplified point system derived here offers a practical tool for real-time decision-making in clinical settings, which can play a crucial role in improving STEMI outcomes.
Figure 1: Representative cases of MIs with and without intramyocardial hemorrhage following reperfusion for STEMI. The top row shows a hemorrhagic MI case, while the bottom row shows a non-hemorrhagic MI case. The first column are pre-PCI angiograms (white arrows point to obstruction), followed by LGE in the second column, highlighting the MI regions, with microvascular obstruction (MVO) appearing as hypointense areas (yellow arrows). The third column shows T2* images, where intramyocardial hemorrhage (IMH) is depicted as hypointense regions (red arrows). 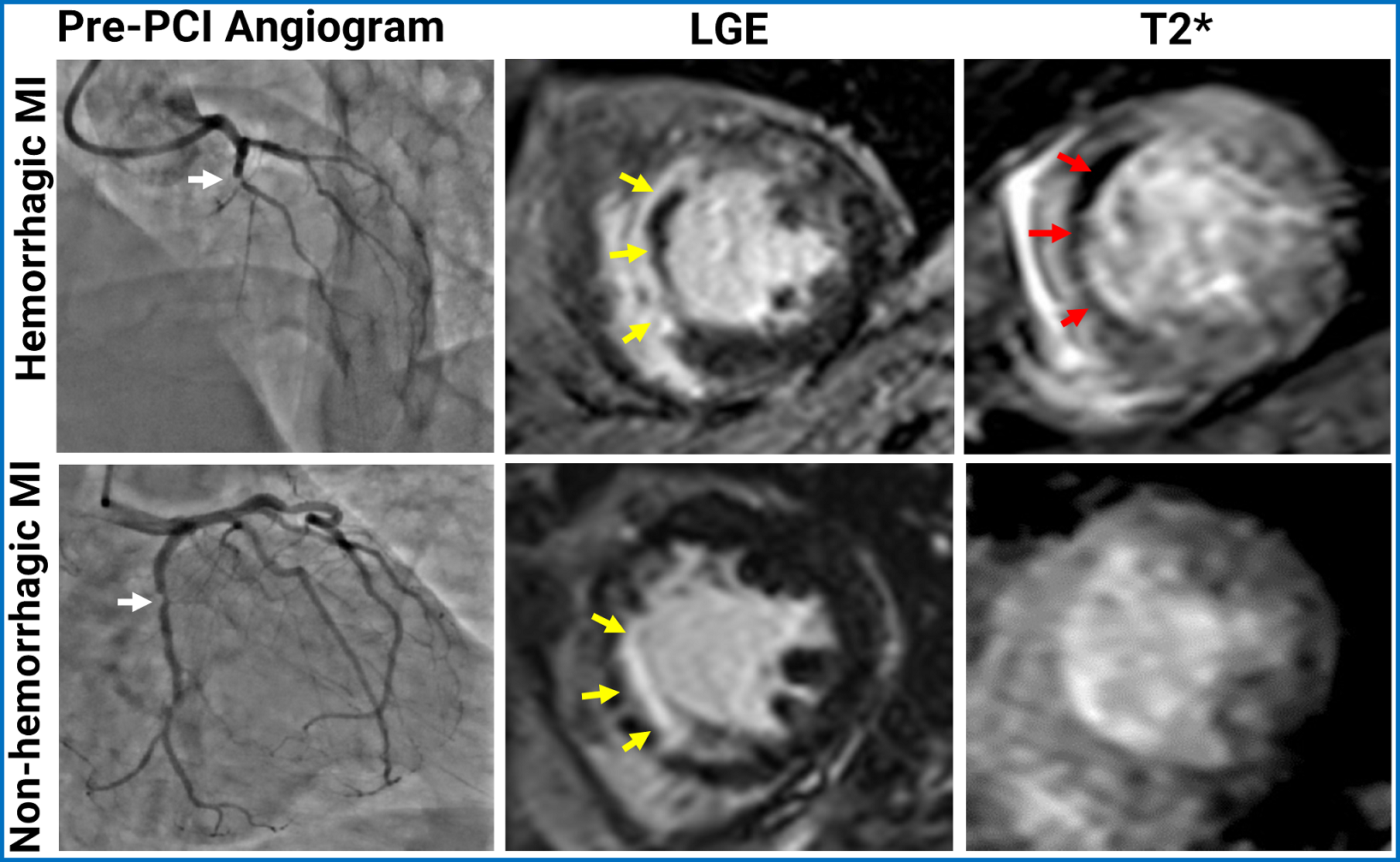
Figure 2: Predictors of IMH Based on Superposable Neural Network (SNN) XAI Algorithm. A total of 12 predictors were included in the analysis (presence of coronary collateral, culprit coronary artery, lesion location, degree of coronary occlusion, gender, age, ST-segment score, BMI, hypertension, diabetes, dyslipidemia, and smoking). Panel (a) shows the relative importance of the individual features as determined by the method. Out of the 12 predictors, the XAI method found collateral presence, and the interaction between ST-segment score and occlusion degree sufficient for obtaining the optimal SNN model. Panel (b) shows the findings from ROC analysis using the SNN model, as well as the threshold used to obtain the point-system as shown in Fig. 3. Panels (c) and (d) show how each predictor in the SNN model relates to probability of observing IMH. Key point values used in creating the point scoring system are highlighted, showing where the threshold can be exceeded by adding the IMH Likelihood from both functions..png)
Figure 3: Predicating IMH with a Point System Derived from the SNN Model. A simplified point system derived from the SNN model offering a practical means for predicting IMH for real-time decision-making in acute care setting. A specified number of points is assigned to each predictor based on its value, and the total number of points determines whether the patient will be hemorrhagic if the total number of points exceeds the specified threshold. For example, a patient with no collaterals, only a partial occlusion, and an ST- segment score of 12 has 1 + 0 + 3 = 4pts in total, thus is expected to develop hemorrhage post reperfusion. In another example, a patient with collaterals, also with a partial occlusion, and an ST score of 8 has 0 + 0 + 2 = 2pts in total, and will not be expected to develop IMH. This point system was found to have an accuracy of 84.9%, sensitivity of 82.3%, and specificity of 87.3%..png)

